Time for some hands-on tutorial on medical imaging. Nevertheless, this time we won’t use loopy AI however primary picture processing algorithms. The objective is to familiarize the reader with ideas round medical imaging and particularly Computed Tomography (CT).
It’s crucial to grasp how far one can go with out deep studying, to grasp when it’s finest to make use of it. New practitioners are likely to ignore that half, however medical picture evaluation continues to be 3D picture processing.
I additionally embrace elements of the code to facilitate the understanding of my thought course of.
The accompanying Google colab pocket book will be discovered right here to run the code proven on this tutorial. A Github repository can be out there. Star our repo for those who appreciated it!
To dive deeper into how AI is utilized in Drugs, you may’t go fallacious with the AI for Drugs on-line course, provided by Coursera. If you wish to concentrate on medical picture evaluation with deep studying, I extremely suggest ranging from the Pytorch-based Udemy Course.
We are going to begin with the very fundamentals of CT imaging. You could skip this part in case you are already aware of CT imaging.
CT imaging
Physics of CT Scans
Computed Tomography (CT) makes use of X-ray beams to acquire 3D pixel intensities of the human physique. A heated cathode releases high-energy beams (electrons), which in flip launch their vitality as X-ray radiation. X-rays cross via human physique tissues and hits a detector on the opposite facet. A dense tissue (i.e. bones) will soak up extra radiation than tender tissues (i.e. fats). When X-rays aren’t absorbed from the physique (i.e. within the air area contained in the lungs) and attain the detector we see them as black, much like a black movie. On the alternative, dense tissues are depicted as white.
On this approach, CT imaging is ready to distinguish density variations and create a 3D picture of the physique.
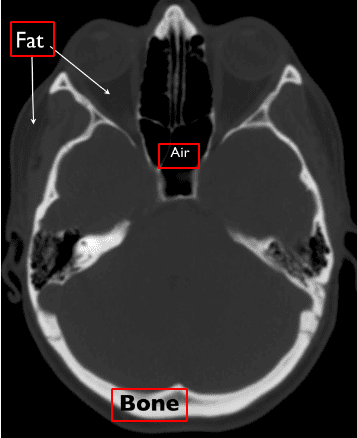
Supply: Christopher P. Hess, M.D., Ph.D, and Derk Purcell, M.D, Division of Radiology and Biomedical Imaging at UCSF
Here’s a 1 min video I discovered very concise:
CT intensities and Hounsfield items
The X-ray absorption is measured within the Hounsfield scale. On this scale, we repair the Air depth to -1000 and water to 0 depth. It’s important to grasp that Housenfield is an absolute scale, not like MRI the place we’ve got a relative scale from 0 to 255.
The picture illustrates a number of the primary tissues and their corresponding depth values. Needless to say the photographs are noisy. The numbers might barely range in actual photographs.
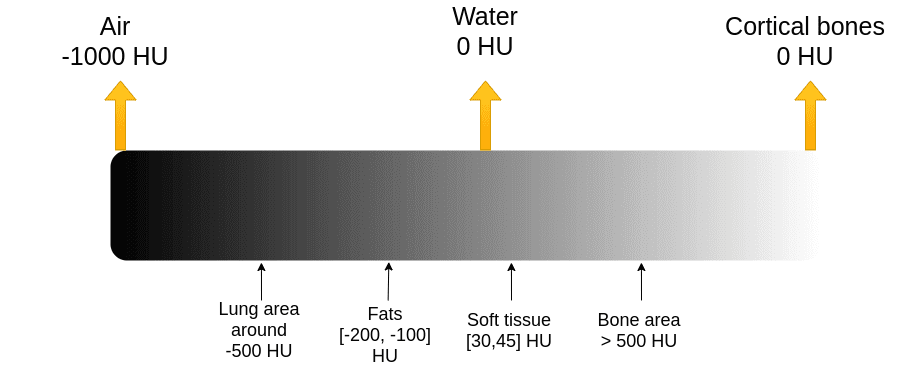
The Hounsfield scale. Picture by Writer.
Bones have excessive depth. We often clip the picture to have an higher most vary. For example, the max worth is perhaps 1000, for sensible causes.
The issue: visualization libraries work on the size [0,255]. It wouldn’t be very smart to visualise all of the Hounsfield scale (from -1000 to 1000+ ) to 256 scales for medical prognosis.
As an alternative, we restrict our consideration to totally different elements of this vary and concentrate on the underlying tissues.
CT knowledge visualization: degree and window
The medical picture conference to clip the Housenfield vary is by selecting a central depth, known as degree and a window, as depicted:
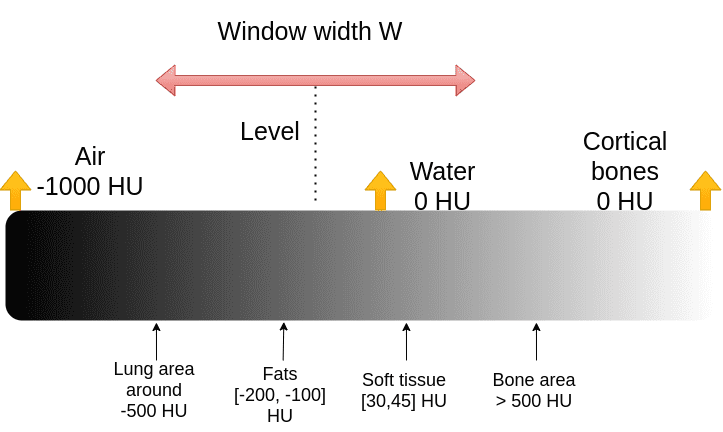
It’s truly fairly an unsightly conference for pc scientists. We’d similar to the min and max of the vary:
import matplotlib.pyplot as plt
import numpy as np
def show_slice_window(slice, degree, window):
"""
Operate to show a picture slice
Enter is a numpy 2D array
"""
max = degree + window/2
min = degree - window/2
slice = slice.clip(min,max)
plt.determine()
plt.imshow(slice.T, cmap="grey", origin="decrease")
plt.savefig('L'+str(degree)+'W'+str(window))
In case you are not satisfied, the following picture will persuade you that the identical CT picture is extra wealthy than a standard picture channel:
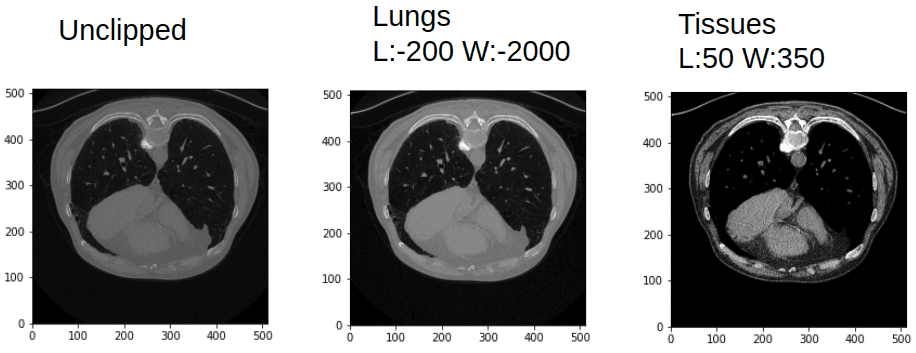
Window and leveling in CT can provide you fairly totally different photographs. Picture by Writer.
For reference here’s a listing of visualization ranges:
| Area/Tissue | Window | Degree |
| mind | 80 | 40 |
| lungs | 1500 | -600 |
| liver | 150 | 30 |
| Tender tissues | 250 | 50 |
| bone | 1800 | 400 |
Time to play!
Lung segmentation based mostly on depth values
We won’t simply phase the lungs however we can even discover the actual space in . To try this we have to discover the actual dimension of the pixel dimensions. Every picture might have a special one (pixdim within the nifty header file). Let’s see the header file first:
import nibabel as nib
ct_img = nib.load(exam_path)
print(ct_img.header)
Right here I’ll present just some vital fields of the header:
<class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
dim : [ 2 512 512 1 1 1 1 1]
datatype : int16
bitpix : 16
pixdim : [1. 0.78515625 0.78515625 1. 1. 1. 1. 1. ]
srow_x : [ -0.78515625 0. 0. 206.60742 ]
srow_y : [ 0. -0.78515625 0. 405.60742 ]
srow_z : [ 0. 0. 1. -304.5]
For the document, srow_x, srow_y, srow_z is the affine matrix of the picture. Bitpix is what number of bits we use to characterize every pixel depth.
So let’s outline a perform that reads that info from the header file. Based mostly on the nifty format every dimension within the nifty file has a pixel dimension. What we’d like is to seek out out the two dimension indices of the picture and their respective pix dimensions.
Step 1: Discover pixel dimensions to calculate the world in mm^2
def find_pix_dim(ct_img):
"""
Get the pixdim of the CT picture.
A common answer that will get the pixdim indicated from the picture dimensions. From the final 2 picture dimensions, we get their pixel dimension.
Args:
ct_img: nib picture
Returns: Checklist of the two pixel dimensions
"""
pix_dim = ct_img.header["pixdim"]
dim = ct_img.header["dim"]
max_indx = np.argmax(dim)
pixdimX = pix_dim[max_indx]
dim = np.delete(dim, max_indx)
pix_dim = np.delete(pix_dim, max_indx)
max_indy = np.argmax(dim)
pixdimY = pix_dim[max_indy]
return [pixdimX, pixdimY]
Step 2: Binarize picture utilizing depth thresholding
We anticipate lungs to be within the Housendfield unit vary of [-1000,-300]. To this finish, we have to clip the picture vary to [-1000,-300] and binarize the values to 0 and 1, so we’ll get one thing like this:
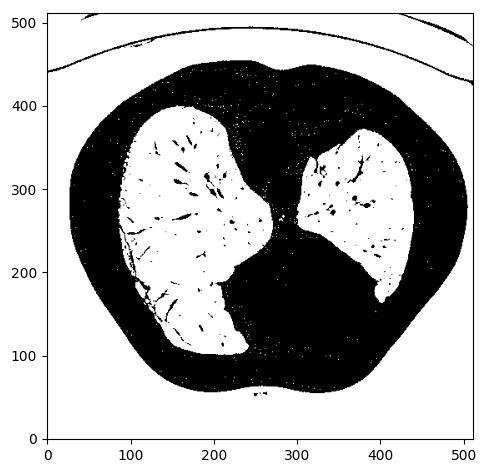
Picture by Writer
Step 3: Contour discovering
Let’s make clear what’s a contour earlier than the rest:
For pc imaginative and prescient, a contour is a set of factors that describe a line or space. So for every detected contour we won’t get a full binary masks however quite a set with a bunch of x and y values.
Okay, how can we isolate the specified space? Hmmm.. let’s give it some thought. We care in regards to the lung areas which might be proven on white. If we might discover an algorithm to establish shut units or any type of contours within the picture which will assist. After some search on-line, I discovered the marching squares technique that finds fixed valued contours in a picture from skimage, known as skimage.measure.find_contours().
After utilizing this perform I visualize the detected contours within the authentic CT picture:
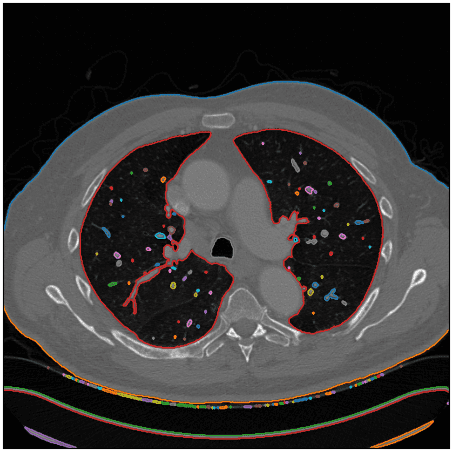
Picture by Writer
Right here is the perform!
def intensity_seg(ct_numpy, min=-1000, max=-300):
clipped = clip_ct(ct_numpy, min, max)
return measure.find_contours(clipped, 0.95)
Step 4: Discover the lung space from a set of attainable contours
Notice that I used a special picture to indicate an edge case that the affected person’s physique will not be a closed set of factors. Okay, not precisely what we would like, however let’s see if we might work that out.
Since we solely care in regards to the lungs we’ve got to set some type of constraints to exclude the undesirable areas.
To take action, I first extracted a convex polygon from the contour utilizing scipy. After I assume 2 constraints:
Which will or might not embrace the physique contour, leading to greater than 3 contours. When that occurs the physique is well discarded by having the most important quantity of the contour that satisfies the pre-described assumptions.
def find_lungs(contours):
"""
Chooses the contours that correspond to the lungs and the physique
First, we exclude non-closed sets-contours
Then we assume some min space and quantity to exclude small contours
Then the physique is excluded as the best quantity closed set
The remaining areas correspond to the lungs
Args:
contours: all of the detected contours
Returns: contours that correspond to the lung space
"""
body_and_lung_contours = []
vol_contours = []
for contour in contours:
hull = ConvexHull(contour)
if hull.quantity > 2000 and set_is_closed(contour):
body_and_lung_contours.append(contour)
vol_contours.append(hull.quantity)
if len(body_and_lung_contours) == 2:
return body_and_lung_contours
elif len(body_and_lung_contours) > 2:
vol_contours, body_and_lung_contours = (listing(t) for t in
zip(*sorted(zip(vol_contours, body_and_lung_contours))))
body_and_lung_contours.pop(-1)
return body_and_lung_contours
As an edge case, I’m displaying that the algorithm will not be restricted to solely two areas of the lungs. Examine the blue contour beneath:
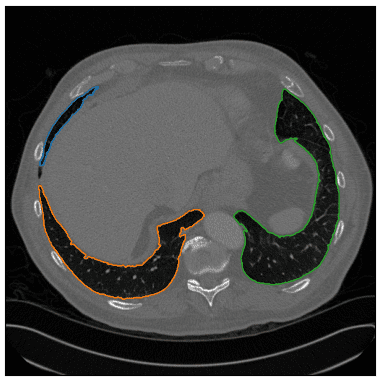
Picture by Writer
Step 5: Contour to binary masks
Subsequent, we put it aside as a nifty file so we have to convert the set of factors to a lung binary masks. For this, I used the pillow python lib that pulls a polygon and creates a binary picture masks. Then I merge all of the masks of the already discovered lung contours.
import numpy as np
from PIL import Picture, ImageDraw
def create_mask_from_polygon(picture, contours):
"""
Creates a binary masks with the size of the picture and
converts the listing of polygon-contours to binary masks and merges them collectively
Args:
picture: the picture that the contours seek advice from
contours: listing of contours
Returns:
"""
lung_mask = np.array(Picture.new('L', picture.form, 0))
for contour in contours:
x = contour[:, 0]
y = contour[:, 1]
polygon_tuple = listing(zip(x, y))
img = Picture.new('L', picture.form, 0)
ImageDraw.Draw(img).polygon(polygon_tuple, define=0, fill=1)
masks = np.array(img)
lung_mask += masks
lung_mask[lung_mask > 1] = 1
return lung_mask.T
The specified lung space in is just the variety of nonzero parts multiplied by the 2 pixel dimensions of the corresponding picture.
The lung areas are saved in a csv file together with the picture identify.
Lastly, to save lots of the masks as nifty I used the worth of 255 for the lung space as an alternative of 1 to have the ability to show in a nifty viewer. Furthermore, I save the picture with the affine transformation of the preliminary CT slice to have the ability to be displayed meaningfully (aligned with none rotation conflicts).
def save_nifty(img_np, identify, affine):
"""
binary masks ought to be transformed to 255 so it may be displayed in a nii viewer
we cross the affine of the preliminary picture to verify it exits in the identical
picture coordinate area
Args:
img_np: the binary masks
identify: output identify
affine: 4x4 np array
Returns:
"""
img_np[img_np == 1] = 255
ni_img = nib.Nifti1Image(img_np, affine)
nib.save(ni_img, identify + '.nii.gz')
Lastly, I opened the masks with a standard nifty viewer for Linux to validate that every part went okay. Listed below are snapshots for slice quantity 4:

Picture by Writer
I used a free medical imaging viewer known as Aliza on Linux.
Phase the principle vessels and compute the vessels over lung space ratio
If there’s a pixel with an depth worth over -500 HU contained in the lung space then we’ll take into account it as a vessel.
First, we do element-wise multiplication between the CT picture and the lung masks to get solely the lungs. Afterwards, we set the zeros that resulted from the element-wise multiplication to -1000 (AIR in HU) and eventually maintain solely the intensities which might be larger than -500 as vessels.
def create_vessel_mask(lung_mask, ct_numpy, denoise=False):
vessels = lung_mask * ct_numpy
vessels[vessels == 0] = -1000
vessels[vessels >= -500] = 1
vessels[vessels < -500] = 0
show_slice(vessels)
if denoise:
return denoise_vessels(lungs_contour, vessels)
show_slice(vessels)
return vessels
One pattern of this course of will be illustrated beneath:
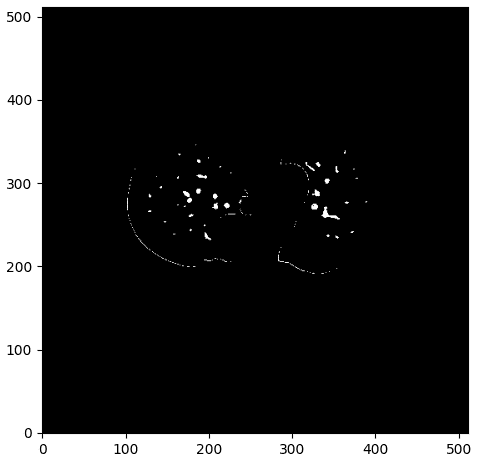
The vessel masks with some noise. Picture by Writer.
Analyzing and enhancing the segmentation’s end result
As you may see we’ve got some elements of the contour of the lungs, which I consider we want to keep away from. To this finish, I created a denoising perform that considers the space of the masks to all of the contour factors. Whether it is beneath 0.1, I set the pixel worth to 0 and because of this exclude them from the detected vessels.
def denoise_vessels(lung_contour, vessels):
vessels_coords_x, vessels_coords_y = np.nonzero(vessels)
for contour in lung_contour:
x_points, y_points = contour[:, 0], contour[:, 1]
for (coord_x, coord_y) in zip(vessels_coords_x, vessels_coords_y):
for (x, y) in zip(x_points, y_points):
d = euclidean_dist(x - coord_x, y - coord_y)
if d <= 0.1:
vessels[coord_x, coord_y] = 0
return vessels
Beneath you may see the distinction between the denoise picture on the proper and the preliminary masks:
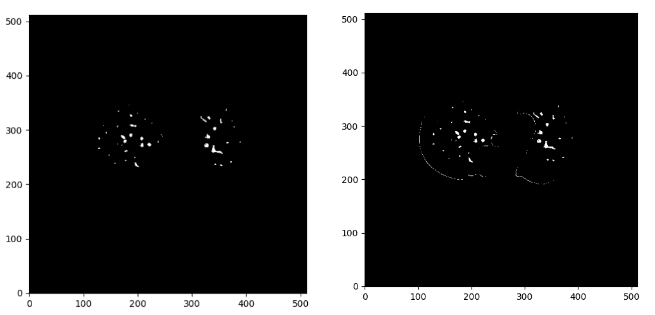
Picture by Writer
If we overlay the masks within the authentic CT picture we get:
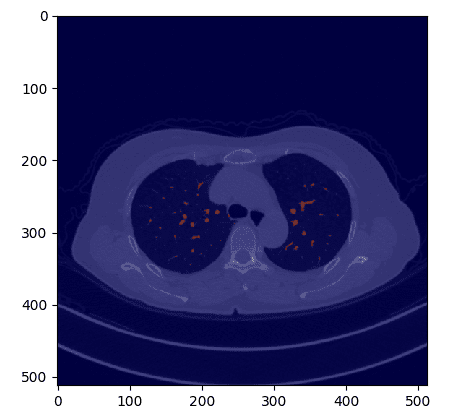
Picture by Writer
def overlay_plot(im, masks):
plt.determine()
plt.imshow(im.T, 'grey', interpolation='none')
plt.imshow(masks.T, 'jet', interpolation='none', alpha=0.5)
Now that we’ve got the masks, the vessel space is computed much like what I did for the lungs, by making an allowance for the person picture pixel dimension.
def compute_area(masks, pixdim):
"""
Computes the world (variety of pixels) of a binary masks and multiplies the pixels
with the pixel dimension of the acquired CT picture
Args:
lung_mask: binary lung masks
pixdim: listing or tuple with two values
Returns: the lung space in mm^2
"""
masks[mask >= 1] = 1
lung_pixels = np.sum(masks)
return lung_pixels * pixdim[0] * pixdim[1]
The ratios are saved in a csv file within the pocket book.
Conclusion and additional readings
I consider that now you could have a strong understanding of CT photographs and their particularities. We are able to do loads of superior issues with such wealthy info of 3D photographs.
Help us by staring our undertaking on GitHub. Lastly, for extra superior tutorials see our medical imaging articles.
I’ve been requested for a big hands-on AI course. I’m considering of writing a e-book on medical imaging in 2021. Till then you may be taught from the coursera course AI for Drugs.
* Disclosure: Please notice that a number of the hyperlinks above is perhaps affiliate hyperlinks, and at no extra value to you, we’ll earn a fee for those who determine to make a purchase order after clicking via.
