A U-shaped structure consists of a selected encoder-decoder scheme: The encoder reduces the spatial dimensions in each layer and will increase the channels. However, the decoder will increase the spatial dims whereas lowering the channels. The tensor that’s handed within the decoder is often referred to as bottleneck. Ultimately, the spatial dims are restored to make a prediction for every pixel within the enter picture. These sorts of fashions are extraordinarily utilized in real-world functions.
This text goals to discover the Unet architectures that stood the check of time.
To dive deeper into how AI is utilized in Medication, you’ll be able to’t go flawed with this on-line course by Coursera: AI for Medication
Absolutely Convolutional Community (FCN)
Absolutely convolutional community 1 was one of many first architectures with out absolutely linked layers. Aside from the truth that it may be skilled end-to-end, for particular person pixel prediction (e.g semantic segmentation), it may possibly course of arbitrary-sized inputs. It’s a normal structure that successfully makes use of transposed convolutions as a trainable upsampling methodology.
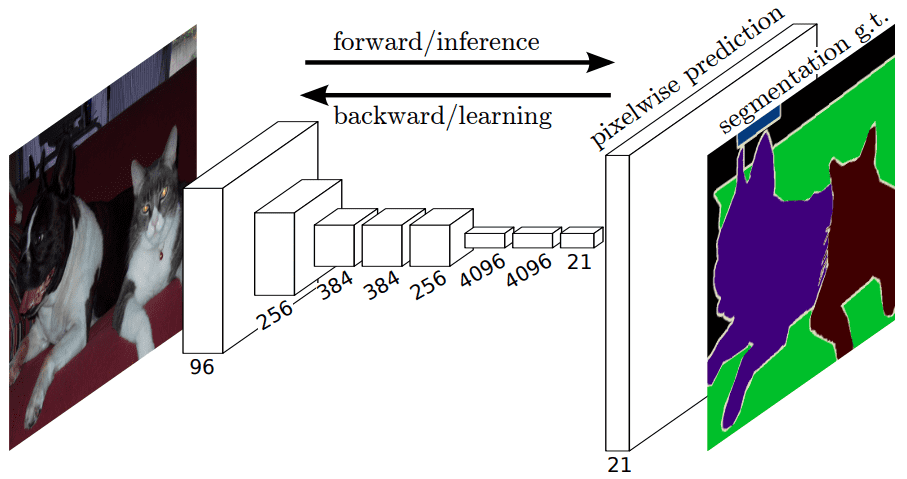
The absolutely convolutional layer structure. Supply
Given a pretrained encoder here’s what an FCN appears like:
import torch
import torch.nn as nn
class FCN32s(nn.Module):
def __init__(self, pretrained_net, n_class):
tremendous().__init__()
self.n_class = n_class
self.pretrained_net = pretrained_net
self.relu = nn.ReLU(inplace=True)
self.deconv1 = nn.ConvTranspose2d(512, 512, kernel_size=3, stride=2, padding=1, dilation=1, output_padding=1)
self.bn1 = nn.BatchNorm2d(512)
self.deconv2 = nn.ConvTranspose2d(512, 256, kernel_size=3, stride=2, padding=1, dilation=1, output_padding=1)
self.bn2 = nn.BatchNorm2d(256)
self.deconv3 = nn.ConvTranspose2d(256, 128, kernel_size=3, stride=2, padding=1, dilation=1, output_padding=1)
self.bn3 = nn.BatchNorm2d(128)
self.deconv4 = nn.ConvTranspose2d(128, 64, kernel_size=3, stride=2, padding=1, dilation=1, output_padding=1)
self.bn4 = nn.BatchNorm2d(64)
self.deconv5 = nn.ConvTranspose2d(64, 32, kernel_size=3, stride=2, padding=1, dilation=1, output_padding=1)
self.bn5 = nn.BatchNorm2d(32)
self.classifier = nn.Conv2d(32, n_class, kernel_size=1)
def ahead(self, x):
output = self.pretrained_net(x)
x5 = output['x5']
rating = self.bn1(self.relu(self.deconv1(x5)))
rating = self.bn2(self.relu(self.deconv2(rating)))
rating = self.bn3(self.relu(self.deconv3(rating)))
rating = self.bn4(self.relu(self.deconv4(rating)))
rating = self.bn5(self.relu(self.deconv5(rating)))
rating = self.classifier(rating)
return rating
You possibly can even load a pretrained mannequin from pytorch hub:
import torch
mannequin = torch.hub.load('pytorch/imaginative and prescient:v0.9.0', 'fcn_resnet101', pretrained=True)
mannequin.eval()
Be aware that every one pre-trained fashions anticipate enter photos normalized in the identical approach, i.e. mini-batches of 3-channel RGB photos of form (N, 3, H, W), the place N is the variety of photos, H and W are anticipated to be not less than 224 pixels. The photographs should be loaded in to a variety of [0, 1] after which normalized utilizing imply = [0.485, 0.456, 0.406] and std = [0.229, 0.224, 0.225]
U-Internet and 3D U-Internet
Afterward, Unet modifies and extends FCN.
The primary thought is to make FCN keep the high-level options within the early layer of the decoder. To this finish, they introduce lengthy skip-connections to localize the segmentations.
On this method, high-resolution options (however semantically low) from the encoder path are mixed and reused with the upsampled output. Unet can also be a symmetric structure, as depicted under.
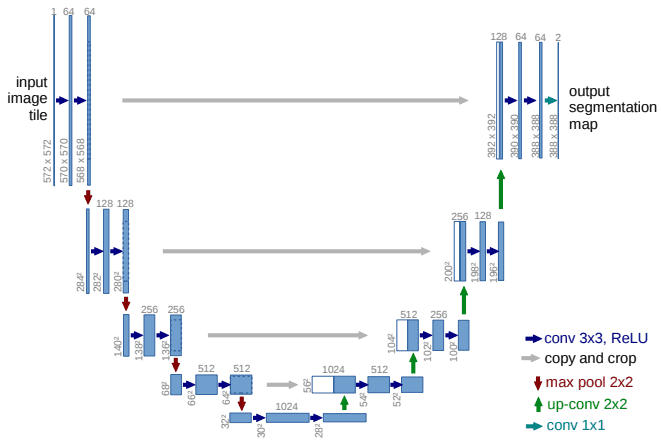
The Unet mannequin. Supply
It may be divided into an encoder-decoder path or contracting-expansive path equivalently.
Encoder (left facet): It consists of the repeated utility of two 3×3 convolutions. Every conv is adopted by a ReLU and batch normalization. Then a 2×2 max pooling operation is utilized to scale back the spatial dimensions. Once more, at every downsampling step, we double the variety of characteristic channels, whereas we lower in half the spatial dimensions.
Decoder path (proper facet): Each step within the expansive path consists of an upsampling of the characteristic map adopted by a 2×2 transpose convolution, which halves the variety of characteristic channels. We even have a concatenation with the corresponding characteristic map from the contracting path, and often a 3×3 convolutional (every adopted by a ReLU). On the last layer, a 1×1 convolution is used to map the channels to the specified variety of lessons.
Right here is an implementation of 2D Unet
import torch
import torch.nn as nn
import torch.nn.useful as F
class DoubleConv(nn.Module):
def __init__(self, in_ch, out_ch):
tremendous(DoubleConv, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True),
nn.Conv2d(out_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True))
def ahead(self, x):
x = self.conv(x)
return x
class InConv(nn.Module):
def __init__(self, in_ch, out_ch):
tremendous(InConv, self).__init__()
self.conv = DoubleConv(in_ch, out_ch)
def ahead(self, x):
x = self.conv(x)
return x
class Down(nn.Module):
def __init__(self, in_ch, out_ch):
tremendous(Down, self).__init__()
self.mpconv = nn.Sequential(
nn.MaxPool2d(2),
DoubleConv(in_ch, out_ch)
)
def ahead(self, x):
x = self.mpconv(x)
return x
class Up(nn.Module):
def __init__(self, in_ch, out_ch, bilinear=True):
tremendous(Up, self).__init__()
if bilinear:
self.up = nn.Upsample(scale_factor=2, mode='bilinear', align_corners=True)
else:
self.up = nn.ConvTranspose2d(in_ch // 2, in_ch // 2, 2, stride=2)
self.conv = DoubleConv(in_ch, out_ch)
def ahead(self, x1, x2):
x1 = self.up(x1)
diffY = x2.dimension()[2] - x1.dimension()[2]
diffX = x2.dimension()[3] - x1.dimension()[3]
x1 = F.pad(x1, (diffX // 2, diffX - diffX // 2,
diffY // 2, diffY - diffY // 2))
x = torch.cat([x2, x1], dim=1)
x = self.conv(x)
return x
class OutConv(nn.Module):
def __init__(self, in_ch, out_ch):
tremendous(OutConv, self).__init__()
self.conv = nn.Conv2d(in_ch, out_ch, 1)
def ahead(self, x):
x = self.conv(x)
return x
class Unet(nn.Module):
def __init__(self, in_channels, lessons):
tremendous(Unet, self).__init__()
self.n_channels = in_channels
self.n_classes = lessons
self.inc = InConv(in_channels, 64)
self.down1 = Down(64, 128)
self.down2 = Down(128, 256)
self.down3 = Down(256, 512)
self.down4 = Down(512, 512)
self.up1 = Up(1024, 256)
self.up2 = Up(512, 128)
self.up3 = Up(256, 64)
self.up4 = Up(128, 64)
self.outc = OutConv(64, lessons)
def ahead(self, x):
x1 = self.inc(x)
x2 = self.down1(x1)
x3 = self.down2(x2)
x4 = self.down3(x3)
x5 = self.down4(x4)
x = self.up1(x5, x4)
x = self.up2(x, x3)
x = self.up3(x, x2)
x = self.up4(x, x1)
x = self.outc(x)
return x
This methodology has nice success in 2D biomedical picture segmentation. And it’s nonetheless used as a baseline methodology. However what about 3D photos?
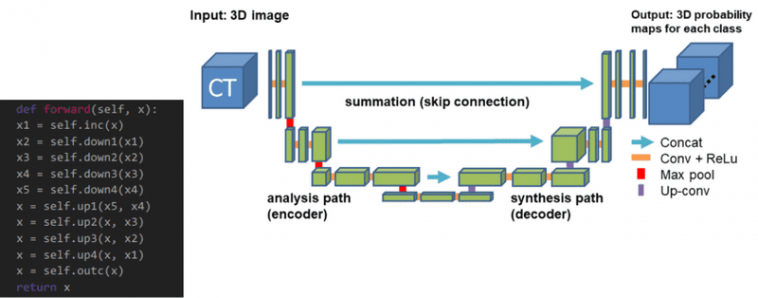
The 3D-Unet
3D Unet was launched shortly after Unet to course of volumes. Solely 3 layers are proven within the official diagram however in apply, we use extra once we implement this mannequin. Every block makes use of batch normalization after the convolution.
The 3D Unet mannequin. Supply
V-Internet (2016)
Vnet extends Unet to course of 3D MRI volumes. In distinction to processing the enter 3D volumes slice-wise, they proposed to make use of 3D convolutions. Ultimately, medical photos have an inherent 3D construction, and slice-wise processing is sub-optimal. The primary modifications of Vnet are:
-
Motivated by related works on picture classification, they changed max-pooling operations with strided convolutions. That is carried out by convolution with 2 × 2 × 2 kernels utilized with stride 2.
-
3D convolutions with padding are carried out in every stage utilizing 5×5×5 kernels.
-
Brief residual connections are additionally employed in each elements of the community.
-
They use 3D transpose convolutions as a way to improve the scale of the inputs, adopted by one to a few conv layers. Characteristic maps are halved in each decoder layer.
All of the above might be illustrated on this picture:
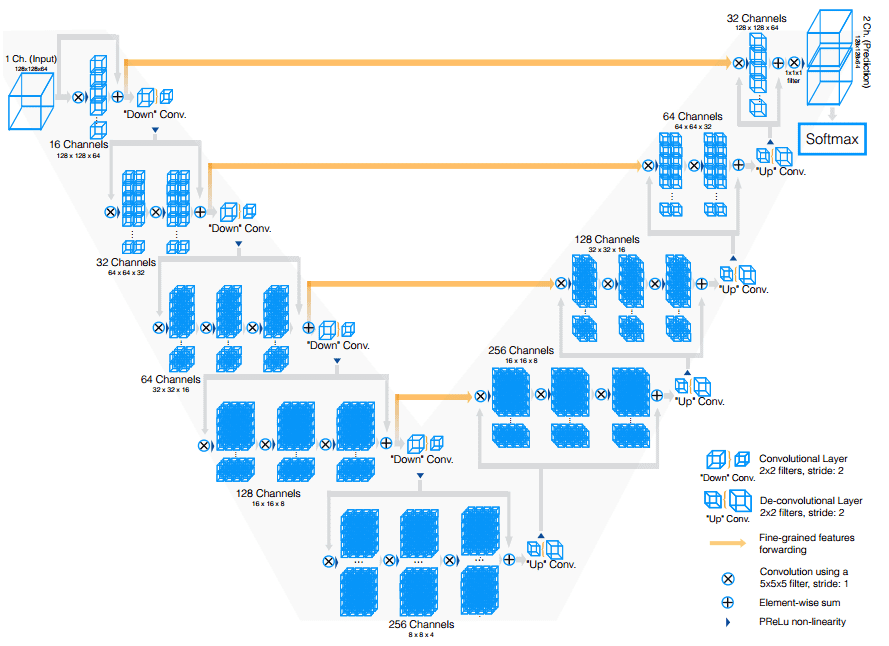
The Vnet mannequin. Supply
Lastly, on this work, the Cube loss was launched which is a standard loss perform in segmentation. You will discover the implementation of Vnet in our open-source library.
UNet++ (2018)
Motivation: The skip connections utilized in U-Internet immediately fast-forward high-resolution characteristic maps from the encoder to the decoder community. This ends in the concatenation of semantically dissimilar characteristic maps.
The primary thought behind UNet++ is to bridge the semantic hole between the characteristic maps of the encoder and decoder earlier than concatenation. To this finish, UNet++ relies on each nested and dense skip connections. UNet++ can successfully seize fine-grained particulars of 2D photos. Visually:
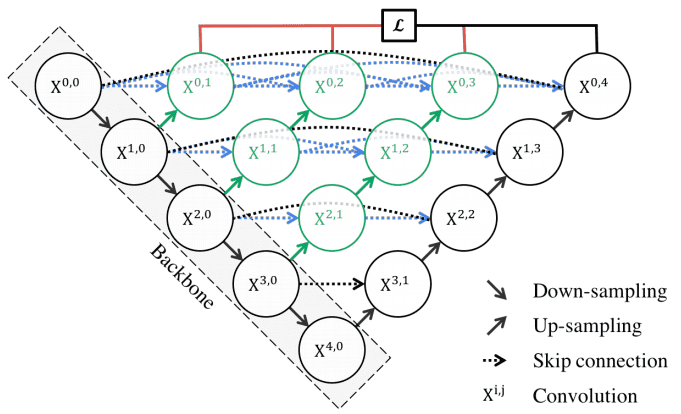
UNet++ consists of an encoder and decoder which are linked by a sequence of nested dense convolutional blocks. Picture by Unet++ paper. Supply
Within the above picture, black signifies the unique U-Internet, whereas inexperienced and blue present dense convolution blocks on the skip pathways. Crimson signifies deep supervision, that means there are a number of loss phrases, versus customary Unet. The implementation is publicly out there.
In different phrases, the dense convolution block brings the semantic stage of the encoder characteristic maps nearer to that of the characteristic maps awaiting within the decoder. Nonetheless, the variety of parameters in addition to the time to coach the community is considerably greater. That is the principle motive that there isn’t a related structure with 3D convolutions.
No New-Internet (2018)
The established Unet baseline for semantic picture segmentation. It was examined on the BRATS dataset with prime ranked outcomes. Details:
-
It makes use of 128x128x128 sub-volumes with a batch dimension of two
-
30 channels within the first conv layer
-
Trilinear upsampling within the decoder
-
Mix Cube loss with adverse log-likelihood
-
Augmentation technique: random rotations, random scaling, random elastic deformations, gamma correction augmentation and mirroring.
-
A l2 weight decay of
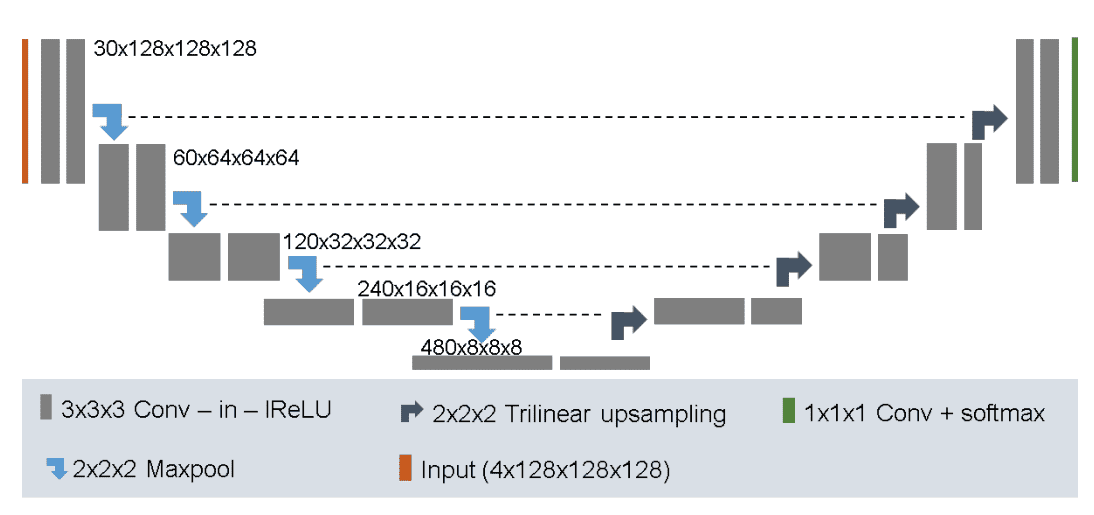
Picture by Fabian Isensee et al. Supply
An in depth walk-through Github repo is on the market.
MRI mind tumor segmentation in 3D utilizing autoencoder regularization
Regardless that this isn’t precisely a standard Unet structure it deserves to belong within the checklist. The encoder is a 3D Resenet mannequin and the decoder makes use of transpose convolutions. The primary essential half is the inexperienced constructing block, as illustrated within the diagram:
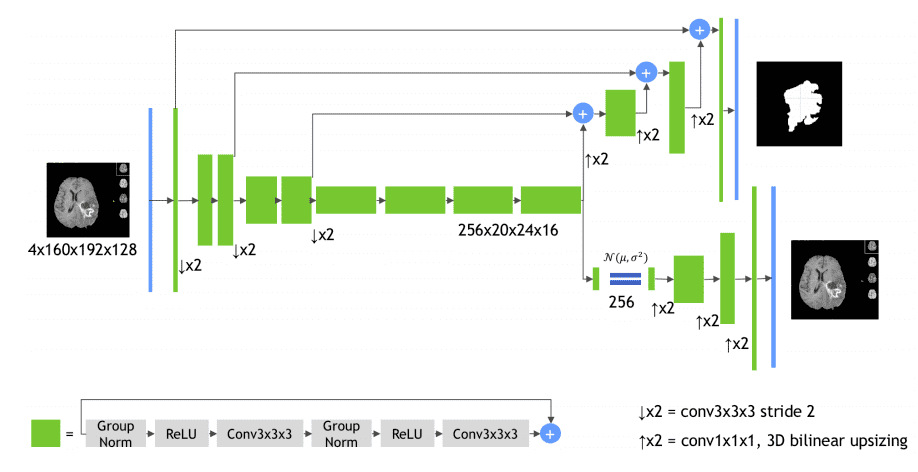
Picture by Andriy Myronenko et al. Supply
It makes use of successive padded 3D convolutions with group normalization, relu activations, and residual skip connections.
The second import part is the subnetwork on the underside proper: it’s a Variational autoencoder that tries to reconstruct the unique 3D enter picture.
Why?
Good query!
The motivation for utilizing the auto-encoder department is to present further steering and regularization to the encoder half, because the coaching information are restricted.
Regardless that this mannequin makes use of an enormous quantity of GPU reminiscence it received the BRATS competitors. Implementation is publicly out there within the MedicalZoo library.
MultiResUNet : Rethinking the U-Internet Structure for Multimodal Biomedical Picture Segmentation (2020)
Medical photos originate from varied modalities and the segmentations we care about are of irregular and completely different scales. To handle this, they proposed to make use of inception-like conv modules. Here’s a fast recap of how the Inception module works:
Following the Inception community, they increase U-Internet with multi-resolutions by incorporating 3 x 3, and seven x 7 convolution operations in parallel to the present 3×3 conv.
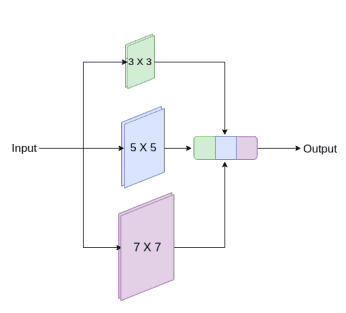
An inception-like block to seize a number of scales. Picture by Nabil Ibtehaz et al. Supply
To cope with the extra community complexity, they factorize the 5 x 5 and seven x 7 convolutional layers, utilizing a sequence of small 3 x 3 convolutional blocks. Then the outputs from the three convolutional blocks are concatenated to extract the spatial options from completely different scales.
Moreover, they progressively improve the filters within the succeeding conv, to scale back the reminiscence footprint of the sooner layers. Lastly, they add a brief residual skip connection and introduce a pointwise (1 x 1) conv layer, which can help us to seize extra spatial data.
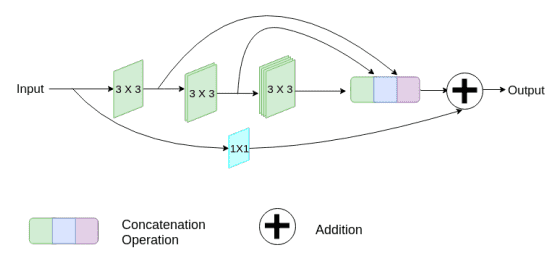
Picture by Nabil Ibtehaz et al. Supply
To deal with the divergence between the encoder-decoder options (because of lengthy skip connections), they suggest to include some convolutional layers alongside the shortcut connections. The speculation is that the extra non-linear transformations ought to compensate the additional processing carried out in the course of the decoder stage.
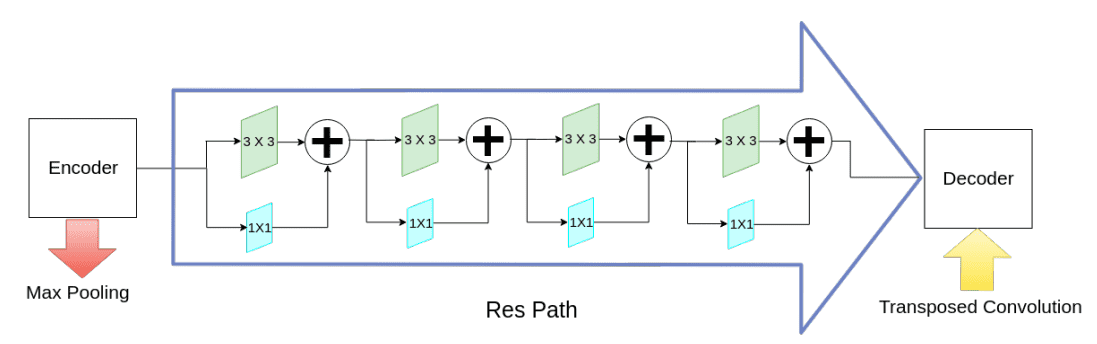
Alternating skip connections. Picture by Nabil Ibtehaz et al. Supply
These architectural inception-like enhancements demonstrated superior ends in many medical picture segmentation datasets. Strive it your self to search out out extra.
The 3D U^2-Internet: introducing channel-wise separable convolutions
Depth-wise implies that the computation is carried out throughout the completely different channels (channel-wise).
In separable convolution, the computation is factorized into two sequential steps: a channel-wise that processes channels independently and one other 1x1xchannel conv that merges the independently produced characteristic maps.
Once more, channel-wise convolution applies an impartial convolutional filter per enter channel,as depicted:
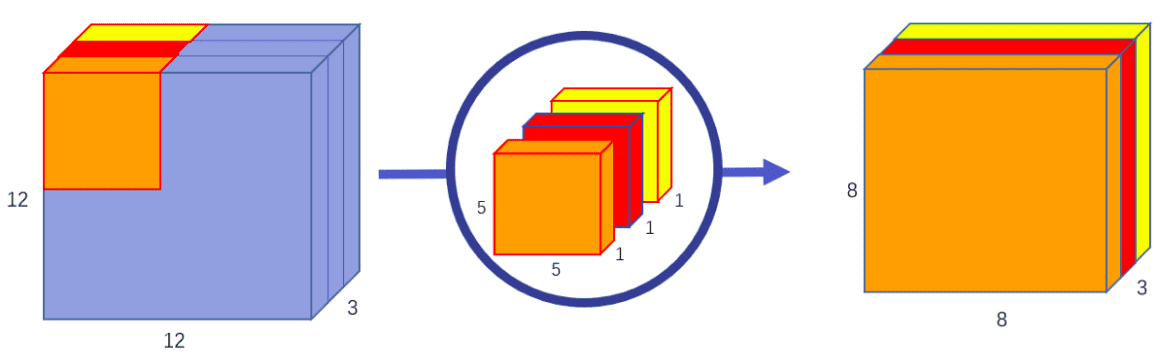
Picture by Chi-Feng Wang. Supply
The pointwise (1x1xk or 1x1x1xk kernel) convolution combines linearly the
output throughout all channels for each spatial location.
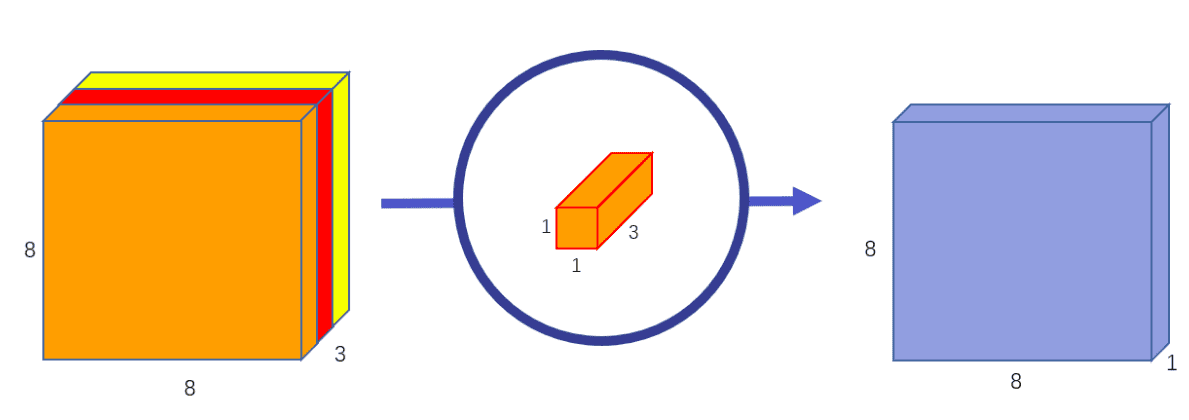
Picture by Chi-Feng Wang. Supply
For the 3D case the parameter achieve is big, particularly when coaching a number of situations of the mannequin on completely different domains, denoted by . A 3x3x3 conv layer with enter channels and out channels want parameters. With separable convolutions the overall variety of weights within the above filter is .
The primary assumption is that every area has its personal channel-wise filters, whereas pointwise conv kernels are shared.
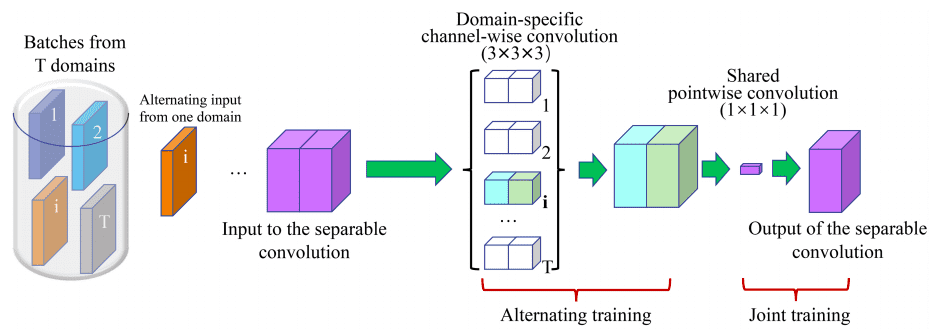
Picture by Chao Huang et al. Supply
The enter layer makes use of 16 filters. The encoder and decoder paths each comprise 5 ranges at completely different resolutions. Residual skip connection is utilized inside every stage. Skip connection is employed to protect extra contextual data from the encoder counterpart for decoder path.
Clearly, the proposed 3D -Internet requires the least parameters, indicating that it may possibly carry out successfully throughout varied domains. The general variety of parameters from the common mannequin is round 1% of that of all impartial fashions, whereas the 2 get hold of comparable segmentation accuracy. Code can also be publicly out there.
Conclusion
To conclude, there isn’t a one dimension suits all mannequin. I attempted to offer a normal set of experimentally validated concepts to work round Unet. Be happy to attempt a few of them out. There may be this git repo that collects Unet architectures with hyperlinks to code. Final however not least, suggestions is at all times welcome! Let me know what you suppose on our social pages.
To dive deeper into how AI is utilized in Medication, we extremely suggest the Coursera course AI for Medication
References
- [1] Lengthy, J., Shelhamer, E., & Darrell, T. (2015). Absolutely convolutional networks for semantic segmentation. In Proceedings of the IEEE convention on laptop imaginative and prescient and sample recognition (pp. 3431-3440).
- [2] Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical picture segmentation. In Worldwide Convention on Medical picture computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
- [3] Milletari, F., Navab, N., & Ahmadi, S. A. (2016, October). V-net: Absolutely convolutional neural networks for volumetric medical picture segmentation. In 2016 fourth worldwide convention on 3D imaginative and prescient (3DV) (pp. 565-571). IEEE.
- [4] Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T., & Ronneberger, O. (2016, October). 3D U-Internet: studying dense volumetric segmentation from sparse annotation. In Worldwide convention on medical picture computing and computer-assisted intervention (pp. 424-432). Springer, Cham.
- [5] Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2018). Unet++: A nested u-net structure for medical picture segmentation. In Deep Studying in Medical Picture Evaluation and Multimodal Studying for Scientific Determination Assist (pp. 3-11). Springer, Cham.
- [6] Wang, W., Yu, Okay., Hugonot, J., Fua, P., & Salzmann, M. (2019). Recurrent U-Internet for resource-constrained segmentation. In Proceedings of the IEEE Worldwide Convention on Pc Imaginative and prescient (pp. 2142-2151).
- [7] Huang, C., Han, H., Yao, Q., Zhu, S., & Zhou, S. Okay. (2019, October). 3D U^2-Internet: A 3D Common U-Internet for Multi-domain Medical Picture Segmentation. In Worldwide Convention on Medical Picture Computing and Pc-Assisted Intervention (pp. 291-299). Springer, Cham.
- [8] Ibtehaz, N., & Rahman, M. S. (2020). MultiResUNet: Rethinking the U-Internet structure for multimodal biomedical picture segmentation. Neural Networks, 121, 74-87.
- [9] Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., & Maier-Hein, Okay. H. (2018, September). No new-net. In Worldwide MICCAI Brainlesion Workshop (pp. 234-244). Springer, Cham.
- [10] Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M., Misawa, Okay., … & Glocker, B. (2018). Consideration u-net: Studying the place to search for the pancreas. arXiv preprint arXiv:1804.03999.
- [11] Alom, M. Z., Hasan, M., Yakopcic, C., Taha, T. M., & Asari, V. Okay. (2018). Recurrent residual convolutional neural community primarily based on u-net (r2u-net) for medical picture segmentation. arXiv preprint arXiv:1802.06955.
- [12] Myronenko, A. (2018, September). 3D MRI mind tumor segmentation utilizing autoencoder regularization. In Worldwide MICCAI Brainlesion Workshop (pp. 311-320). Springer, Cham.
* Disclosure: Please word that among the hyperlinks above is likely to be affiliate hyperlinks, and at no extra value to you, we are going to earn a fee for those who resolve to make a purchase order after clicking by.