Once I realized that I can not apply frequent picture processing pipelines in medical photographs, I used to be fully discouraged. Why does such performance not exist? So, I made up this put up (plus a pocket book) for discouraged people who, like me, are involved in fixing medical imaging issues.
We’ve already mentioned medical picture segmentation and a few preliminary background on coordinate techniques and DICOM recordsdata. My expertise within the discipline leads me to proceed with knowledge understanding, preprocessing, and a few augmentations. As I all the time say, if you happen to merely perceive your knowledge and their particularities, you might be most likely enjoying bingo. Within the discipline of medical imaging, I discover some knowledge manipulations, that are closely utilized in preprocessing and augmentation in state-of-the-art strategies, to be vital in our understanding. To this finish, I present a pocket book for everybody to mess around. It performs transformations on medical photographs, which is just a 3D structured grid.
To dive deeper into how AI is utilized in Drugs, you possibly can’t go flawed with the AI for Drugs on-line course, supplied by Coursera. If you wish to deal with medical picture evaluation with deep studying, I extremely suggest ranging from the Pytorch-based Udemy Course.
Information: We are going to play with 2 MRI photographs which are supplied from nibabel (python library) for illustration functions. The photographs are saved as nifty recordsdata. However earlier than that, let’s write up some code to visualise the 3D medical volumes.
The photographs might be proven in 3 planes: sagittal, coronal, axial trying from left to proper all through this put up.
Two-dimensional planes visualization
All through the entire tutorial, we’ll extensively use a operate that visualizes the three median slices within the sagittal, coronal, and axial planes respectively. Let’s write some minimal operate to take action:
def show_mid_slice(img_numpy, title='img'):
"""
Accepts an 3D numpy array and reveals median slices in all three planes
"""
assert img_numpy.ndim == 3
n_i, n_j, n_k = img_numpy.form
center_i1 = int((n_i - 1) / 2)
center_j1 = int((n_j - 1) / 2)
center_k1 = int((n_k - 1) / 2)
show_slices([img_numpy[center_i1, :, :],
img_numpy[:, center_j1, :],
img_numpy[:, :, center_k1]])
plt.suptitle(title)
def show_slices(slices):
"""
Operate to show a row of picture slices
Enter is a listing of numpy 2D picture slices
"""
fig, axes = plt.subplots(1, len(slices))
for i, slice in enumerate(slices):
axes[i].imshow(slice.T, cmap="grey", origin="decrease")
Nothing greater than matplotlib’s “imshow" and numpy’s array manipulations. For the report, medical photographs are a single channel and we visualize them in grayscale colours.
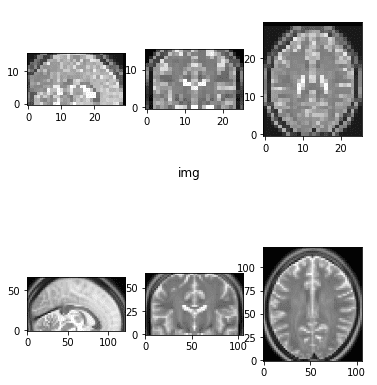
The 2 photographs that we’ll use to play with a plethora of transformations might be illustrated beneath:
show_mid_slice(epi_img_numpy, 'first picture')
show_mid_slice(anatomy_img_numpy,'second picture')
The preliminary mind MRI photographs that we’ll use.
Now we’re good to go! Let’s start with resize and rescale in medical photographs. Yeap, it’s not precisely the identical.
Medical picture resizing (down/up-sampling)
The scipy library offers numerous functionalities for multi-dimensional photographs. Since medical photographs are three dimensional, numerous functionalities can be utilized. This time we’ll use scipy.ndimage.interpolation.zoom for resizing the picture within the desired dimensions. That is much like downsampling in a 2D picture. The identical operate can be utilized for interpolation to extend the spatial dimensions. As an illustration, we’ll double and half the unique picture dimension.
Remember that in this sort of transformation the ratios are normally essential to be maintained.
You most likely don’t wish to lose the anatomy of the human physique 🙂
import scipy
def resize_data_volume_by_scale(knowledge, scale):
"""
Resize the information based mostly on the supplied scale
"""
scale_list = [scale,scale,scale]
return scipy.ndimage.interpolation.zoom(knowledge, scale_list, order=0)
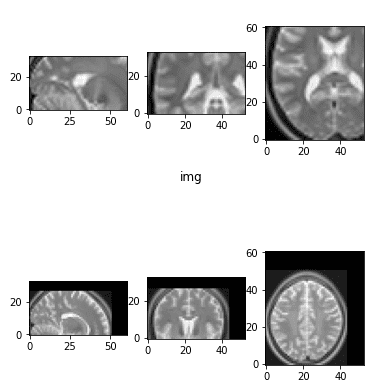
end result = resize_data_volume_by_scale(epi_img_numpy, 0.5)
result2 = resize_data_volume_by_scale(epi_img_numpy, 2)
This sort of scaling is normally known as isometric. Truthfully, I’m not an enormous fan of the scipy’s terminology to make use of the phrase zoom for this performance.
Downsampled and upsampled picture by an element of two
It is vitally frequent to downsample the picture in a decrease dimension for heavy machine studying.
Word that there’s one other sort of resizing. As a substitute of offering the specified output form, you specify the specified voxel dimension(i.e. voxel_size=(1,1,1) mm). Nibabel offers a operate known as resample_to_output(). It really works with nifti recordsdata and never with numpy arrays. Truthfully, I would not suggest it alone because the ensuing photographs may not have the identical form. This can be an issue for deep studying. For instance to create batches with dataloaders the dimension ought to be constant throughout situations. Nevertheless, chances are you’ll select to incorporate it in a earlier step in your pipeline. It may be used to deliver totally different photographs to have the identical or comparable voxel dimension.
Medical picture rescaling (zoom- in/out)
Rescaling might be considered an affine transformation. We are going to randomly zoom out and in of the picture. This augmentation normally helps the mannequin to be taught scale-invariant options.
def random_zoom(matrix,min_percentage=0.7, max_percentage=1.2):
z = np.random.pattern() *(max_percentage-min_percentage) + min_percentage
zoom_matrix = np.array([[z, 0, 0, 0],
[0, z, 0, 0],
[0, 0, z, 0],
[0, 0, 0, 1]])
return ndimage.interpolation.affine_transform(matrix, zoom_matrix)
Random zoom in and zoom out
You will need to see that the empty space is stuffed with black pixels (zero depth).
Word right here that the encircling air in medical photographs doesn’t have zero depth. Black is actually relative to medical photographs.
The following one on the record is 3D rotation.
Medical picture rotation
Rotation is among the most typical strategies to realize knowledge augmentation in laptop imaginative and prescient. It has additionally been thought-about a self-supervised method with exceptional outcomes [Spyros Gidaris et al. ].
In medical imaging, it’s an equal import performance that has additionally been used from self-supervised pretraining [Xinrui Zhuang et al. 2019 ]. A easy random 3D rotation in a given vary of levels might be illustrated with the code beneath:
def random_rotate3D(img_numpy, min_angle, max_angle):
"""
Returns a random rotated array in the identical form
:param img_numpy: 3D numpy array
:param min_angle: in levels
:param max_angle: in levels
"""
assert img_numpy.ndim == 3, "present a 3d numpy array"
assert min_angle < max_angle, "min ought to be lower than max val"
assert min_angle > -360 or max_angle < 360
all_axes = [(1, 0), (1, 2), (0, 2)]
angle = np.random.randint(low=min_angle, excessive=max_angle+1)
axes_random_id = np.random.randint(low=0, excessive=len(all_axes))
axes = all_axes[axes_random_id]
return scipy.ndimage.rotate(img_numpy, angle, axes=axes)
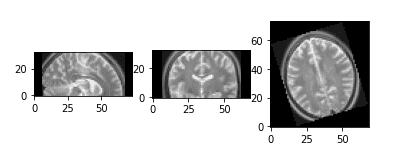
Random 3D rotation
We merely need to outline the axis and the rotation angle. As scaling supplied the mannequin with extra variety with a view to be taught scale-invariant options, rotation aids in studying rotation-invariant options.
Subsequent in our record is picture flipping.
Medical picture flip
Just like frequent RGB photographs, we will carry out axis flipping in medical photographs. At this level, it’s actually essential to make clear one factor:
After we carry out augmentations and/or preprocessing in our knowledge, we could have to use comparable operations on the bottom reality knowledge.
As an illustration, if we deal with the duty of medical picture segmentation, you will need to flip the goal segmentation map. A easy implementation might be discovered beneath:
def random_flip(img, label=None):
axes = [0, 1, 2]
rand = np.random.randint(0, 3)
img = flip_axis(img, axes[rand])
img = np.squeeze(img)
if label is None:
return img
else:
y = flip_axis(y, axes[rand])
y = np.squeeze(y)
return x, y
def flip_axis(x, axis):
x = np.asarray(x).swapaxes(axis, 0)
x = x[::-1, ...]
x = x.swapaxes(0, axis)
return x
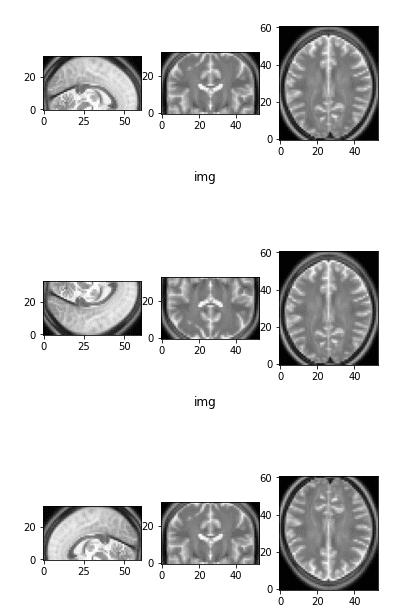
The preliminary picture as a reference and two flipped variations
Observe that by flipping one axis, two views change. The primary picture on prime is the preliminary picture as a reference.
Medical picture shifting (displacement)
Right here I wish to inform one thing else.
Rotation, shifting, and scaling are nothing greater than affine transformations.
Generally I implement them by simply defining the affine transformations and apply it within the picture with scipy, and generally I take advantage of the already-implemented capabilities for multi-dimensional picture processing.
So as to use this operation in my knowledge augmentation pipeline, you possibly can see that I’ve included a wrapper operate. The latter principally samples a random quantity, normally within the desired vary, and calls the affine transformation operate. Under is the implementation for random shifting/displacement.
def transform_matrix_offset_center_3d(matrix, x, y, z):
offset_matrix = np.array([[1, 0, 0, x], [0, 1, 0, y], [0, 0, 1, z], [0, 0, 0, 1]])
return ndimage.interpolation.affine_transform(matrix, offset_matrix)
def random_shift(img_numpy, max_percentage=0.4):
dim1, dim2, dim3 = img_numpy.form
m1,m2,m3 = int(dim1*max_percentage/2),int(dim1*max_percentage/2), int(dim1*max_percentage/2)
d1 = np.random.randint(-m1,m1)
d2 = np.random.randint(-m2,m2)
d3 = np.random.randint(-m3,m3)
return transform_matrix_offset_center_3d(img_numpy, d1, d2, d3)
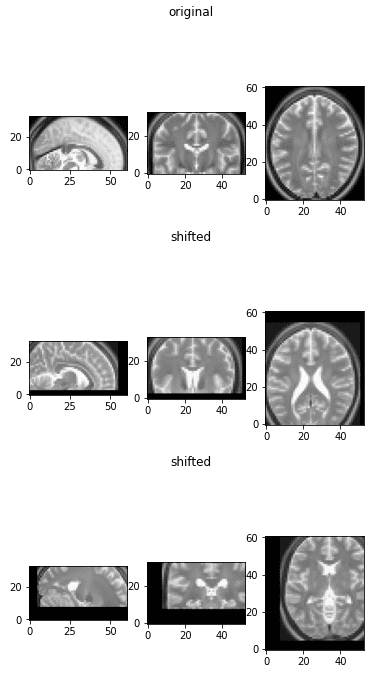
The displaced medical photographs
This augmentation isn’t quite common in medical picture augmentation, however we embody them right here for completeness.
The explanation we don’t embody it’s that convolutional neural networks are by definition designed to be taught translation-invariant options.
Random 3D crop
Cropping isn’t considerably totally different from pure photographs additionally. Nevertheless, remember the fact that we normally need to take all of the slices of a dimension and we have to handle that. The reason being that one dimension could have fewer slices than the others. For instance, one time I needed to take care of a 384x384x64 picture, which is frequent in CT photographs.
def crop_3d_volume(img_tensor, crop_dim, crop_size):
assert img_tensor.ndim == 3, '3d tensor have to be supplied'
full_dim1, full_dim2, full_dim3 = img_tensor.form
slices_crop, w_crop, h_crop = crop_dim
dim1, dim2, dim3 = crop_size
if full_dim1 == dim1:
img_tensor = img_tensor[:, w_crop:w_crop + dim2,
h_crop:h_crop + dim3]
elif full_dim2 == dim2:
img_tensor = img_tensor[slices_crop:slices_crop + dim1, :,
h_crop:h_crop + dim3]
elif full_dim3 == dim3:
img_tensor = img_tensor[slices_crop:slices_crop + dim1, w_crop:w_crop + dim2, :]
else:
img_tensor = img_tensor[slices_crop:slices_crop + dim1, w_crop:w_crop + dim2,
h_crop:h_crop + dim3]
return img_tensor
def find_random_crop_dim(full_vol_dim, crop_size):
assert full_vol_dim[0] >= crop_size[0], "crop dimension is simply too large"
assert full_vol_dim[1] >= crop_size[1], "crop dimension is simply too large"
assert full_vol_dim[2] >= crop_size[2], "crop dimension is simply too large"
if full_vol_dim[0] == crop_size[0]:
slices = crop_size[0]
else:
slices = np.random.randint(full_vol_dim[0] - crop_size[0])
if full_vol_dim[1] == crop_size[1]:
w_crop = crop_size[1]
else:
w_crop = np.random.randint(full_vol_dim[1] - crop_size[1])
if full_vol_dim[2] == crop_size[2]:
h_crop = crop_size[2]
else:
h_crop = np.random.randint(full_vol_dim[2] - crop_size[2])
return (slices, w_crop, h_crop)
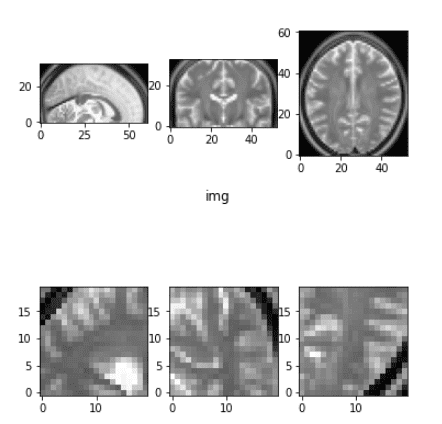
Random cropping instance
There are different methods for cropping that target the world that we have an interest i.e. the tumor, however we is not going to get into that now.
Thus far we performed with geometrical transformations. Let’s see what we will do with the depth of the picture.
Clip depth values (outliers)
This step isn’t relevant for this tutorial, however it could are available in fairly helpful usually. Particularly for CT photographs. The explanation it’s not relevant is that the MRI photographs are in a fairly slim vary of values.
def percentile_clip(img_numpy, min_val=5, max_val=95):
"""
Depth normalization based mostly on percentile
Clips the vary based mostly on the quartile values.
:param min_val: ought to be within the vary [0,100]
:param max_val: ought to be within the vary [0,100]
:return: depth normalized picture
"""
low = np.percentile(img_numpy, min_val)
excessive = np.percentile(img_numpy, max_val)
img_numpy[img_numpy < low] = low
img_numpy[img_numpy > high] = excessive
return img_numpy
def clip_range(img_numpy, min_intensity=10, max_intensity=240):
return np.clip(img_numpy, min_intensity, max_intensity)
Depth normalization in medical photographs
Right here, I embody the most typical depth normalizations: min-max and imply/std. One little factor to bear in mind:
After we carry out imply/std normalization we normally omit the zero depth voxels from the calculation of the imply. This holds true largely for MRI photographs.
A technique to have a look at that is if we’ve got a mind picture; we most likely do not wish to normalize it with the depth of the voxels round it.
def normalize_intensity(img_tensor, normalization="imply"):
"""
Accepts a picture tensor and normalizes it
:param normalization: selections = "max", "imply" , sort=str
For imply normalization we use the non zero voxels solely.
"""
if normalization == "imply":
masks = img_tensor.ne(0.0)
desired = img_tensor[mask]
mean_val, std_val = desired.imply(), desired.std()
img_tensor = (img_tensor - mean_val) / std_val
elif normalization == "max":
MAX, MIN = img_tensor.max(), img_tensor.min()
img_tensor = (img_tensor - MIN) / (MAX - MIN)
return img_tensor
There isn’t a level to visualise this transformation as its goal is to feed the preprocessed knowledge into the deep studying mannequin. In fact, some other form of depth normalization could apply in medical photographs.
Elastic deformation
Once I first learn this transformation within the unique Unet paper, I didn’t perceive a single phrase from the paragraph:
“As for our duties there’s little or no coaching knowledge obtainable, we use extreme knowledge augmentation by making use of elastic deformations to the obtainable coaching photographs. This permits the community to be taught invariance to such deformations, with out the necessity to see these transformations within the annotated picture corpus. That is significantly essential in biomedical segmentation since deformation was the most typical variation in tissue and sensible deformations might be simulated effectively” ~ Olaf Ronneberger et al. 2015 (Unet paper)
Truthfully, I haven’t seemed into the unique publication of 2003. And also you most likely gained’t additionally. I seemed into another code implementations and tried to make it extra easy. I made a decision to incorporate it in my tutorial as a result of you will notice it so much in literature.
def elastic_transform_3d(picture, labels=None, alpha=4, sigma=35, bg_val=0.1):
"""
Elastic deformation of photographs as described in
Simard, Steinkraus and Platt, "Finest Practices for
Convolutional Neural Networks utilized to Visible
Doc Evaluation", in
Proc. of the Worldwide Convention on Doc Evaluation and
Recognition, 2003.
Modified from:
https://gist.github.com/chsasank/4d8f68caf01f041a6453e67fb30f8f5a
https://github.com/fcalvet/image_tools/blob/grasp/image_augmentation.py#L62
Modified to take 3D inputs
Deforms each the picture and corresponding label file
picture linear/trilinear interpolated
Label volumes nearest neighbour interpolated
"""
assert picture.ndim == 3
form = picture.form
dtype = picture.dtype
coords = np.arange(form[0]), np.arange(form[1]), np.arange(form[2])
im_intrps = RegularGridInterpolator(coords, picture,
methodology="linear",
bounds_error=False,
fill_value=bg_val)
dx = gaussian_filter((np.random.rand(*form) * 2 - 1), sigma,
mode="fixed", cval=0.) * alpha
dy = gaussian_filter((np.random.rand(*form) * 2 - 1), sigma,
mode="fixed", cval=0.) * alpha
dz = gaussian_filter((np.random.rand(*form) * 2 - 1), sigma,
mode="fixed", cval=0.) * alpha
x, y, z = np.mgrid[0:shape[0], 0:form[1], 0:form[2]]
indices = np.reshape(x + dx, (-1, 1)),
np.reshape(y + dy, (-1, 1)),
np.reshape(z + dz, (-1, 1))
picture = np.empty(form=picture.form, dtype=dtype)
picture = im_intrps(indices).reshape(form)
if labels is not None:
lab_intrp = RegularGridInterpolator(coords, labels,
methodology="nearest",
bounds_error=False,
fill_value=0)
labels = lab_intrp(indices).reshape(form).astype(labels.dtype)
return picture, labels
return picture
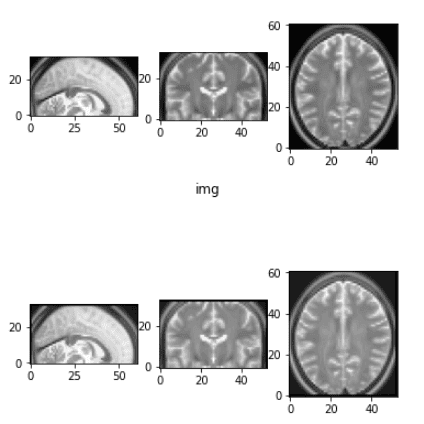
Earlier than and after the elastic deformation
What it is advisable to take into consideration is that this transformation adjustments the depth and applies some Gaussian noise in every dimension. For extra data it’s important to get again to the unique work.
Conclusion
By now you possibly can resonate with my ideas on the particularities on medical imaging preprocessing and augmentations. However don’t neglect: you possibly can play with the tutorial on-line and see the transformations by your self. It helps, consider me. Understanding our medical photographs is essential. Now you can select which transformations to use in your undertaking.
In case you appreciated our tutorial, please be at liberty to share it in your social media web page, as a reward for our work. It might be extremely appreciated.
Keep tuned for extra AI summer time articles!
* Disclosure: Please be aware that a number of the hyperlinks above could be affiliate hyperlinks, and at no extra price to you, we’ll earn a fee if you happen to resolve to make a purchase order after clicking by means of.


